Polymerase-DNA
Interactions
About Klenow and Klentaq
E. coli ‘Pol I’ and T. aquaticus’ Taq polymerase’ are homologous DNA polymerase I proteins which display high sequence, morphological and functional similarities. Klenow, the “large fragment domain” of E. coli Pol I is a mesophilic protein, and denatures at around 40°C. Klentaq, the “large fragment domain” of the Taq polymerase is a thermophilic protein and is active and stable up to 100°C. Morphologically, the proteins are often described as a half-open open right hand with the polymerase active site at the palm and the proofreading domain at the “heel” of the hand.
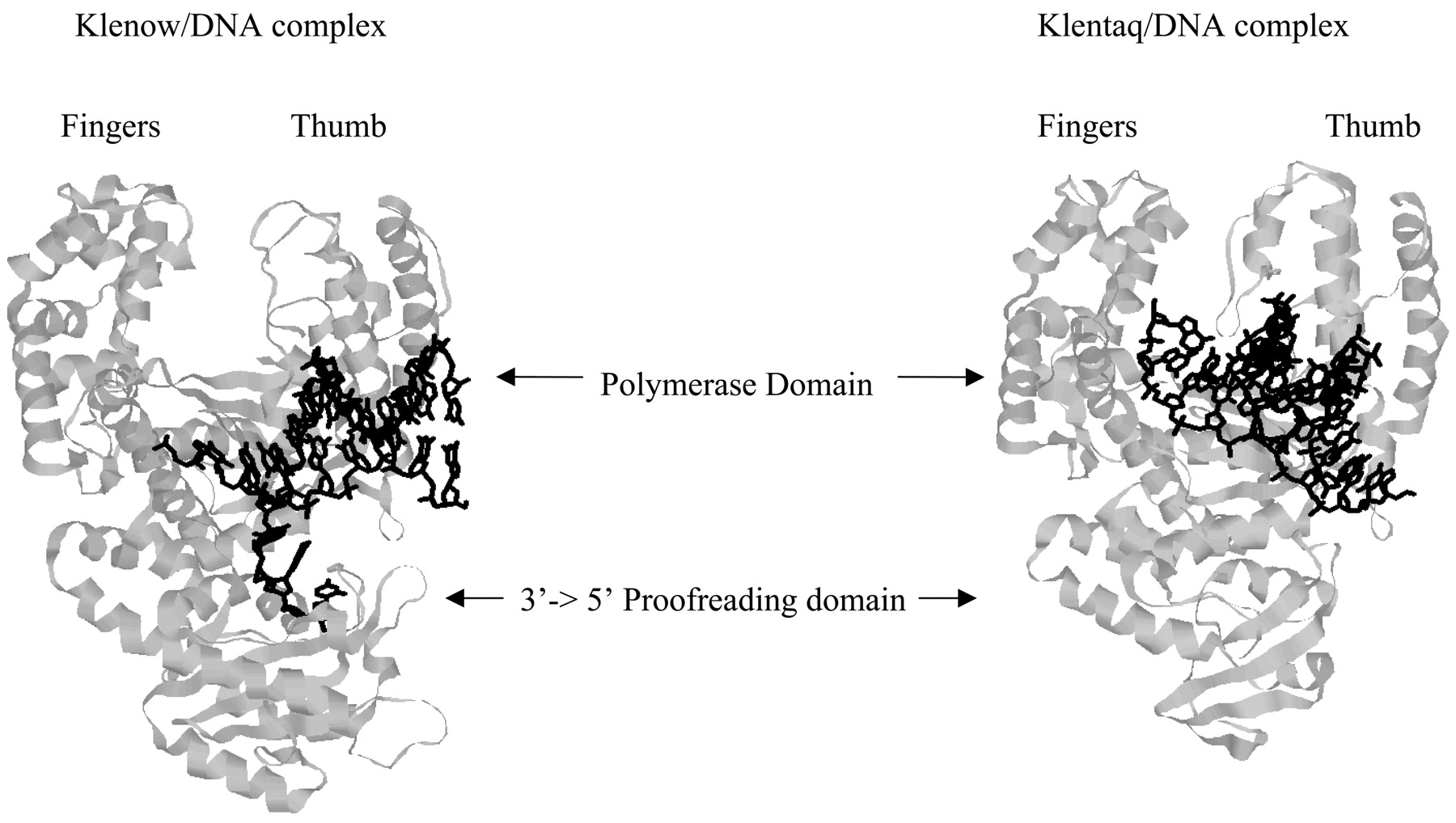
Figure 1. Structure of Klenow/DNA complex (1KLN) and Klentaq/DNA complex (4KTQ). Klenow and Klentaq have homologous structures. Klenow possesses 3' - 5' proofreading activity while Klentaq does not.
Salt Dependence of Binding
The environment (e.g. salt concentration and type, pH, temperature, etc.) exerts large effects on the DNA binding by these polymerases. We have determined the salt dependence of DNA binding for both Klenow and Klentaq polymerases. In Figure 2, below, as the concentration of KCl is increase, the binding curves shift to the right, indicating weaker binding with added salt.
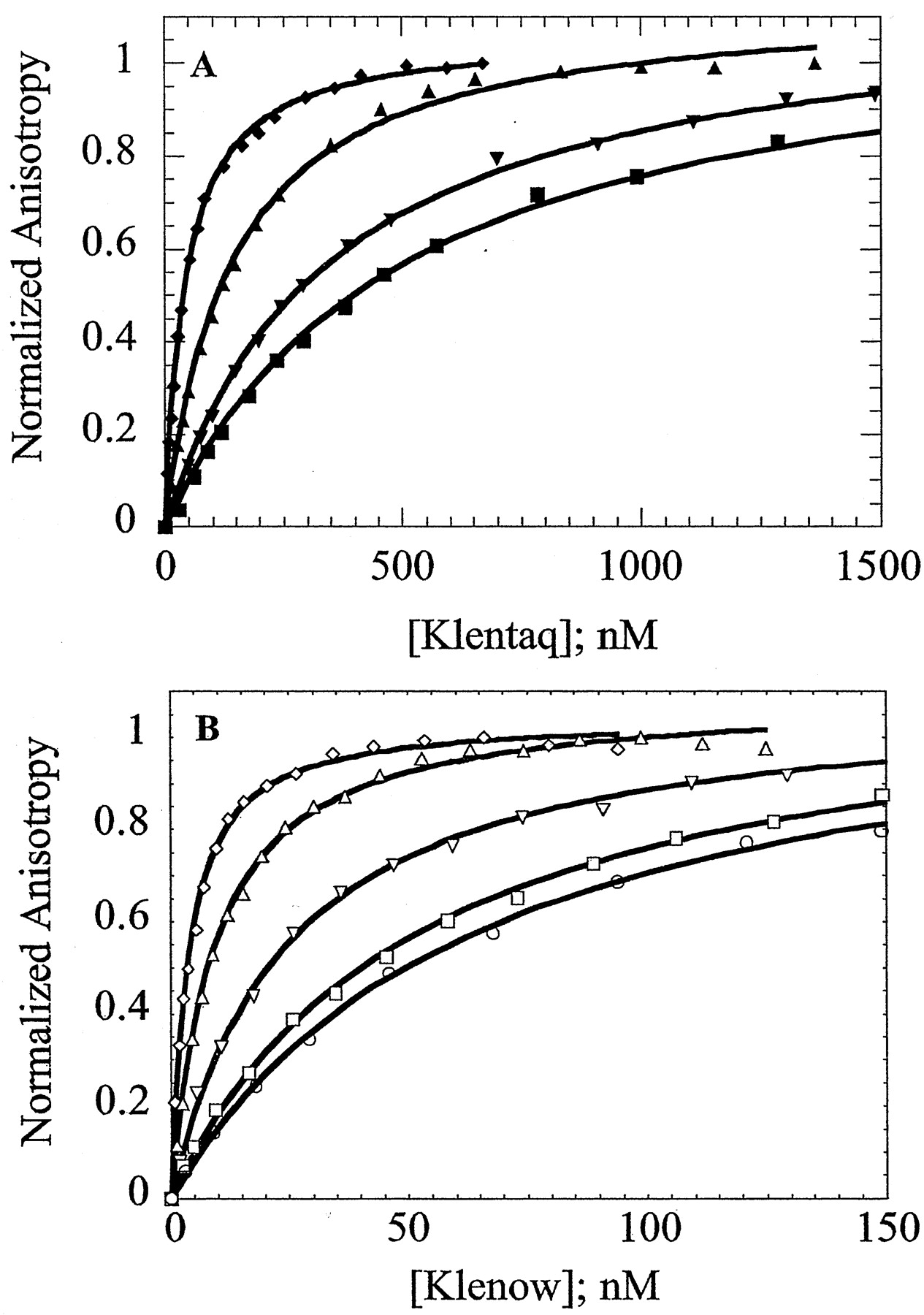
Figure 2. Salt dependence of primed-template DNA binding by Klenow and Klentaq . Panel A = equilibrium titrations of Klentaq at KCl concentrations of 75 mM, 125 mM, 150 mM, and 175 mM. Panel B = equilibrium titrations of Klenow at KCl concentrations of 250 mM, 300 mM, 400 mM, 450 mM, and 500 mM. (from Ref. 1).
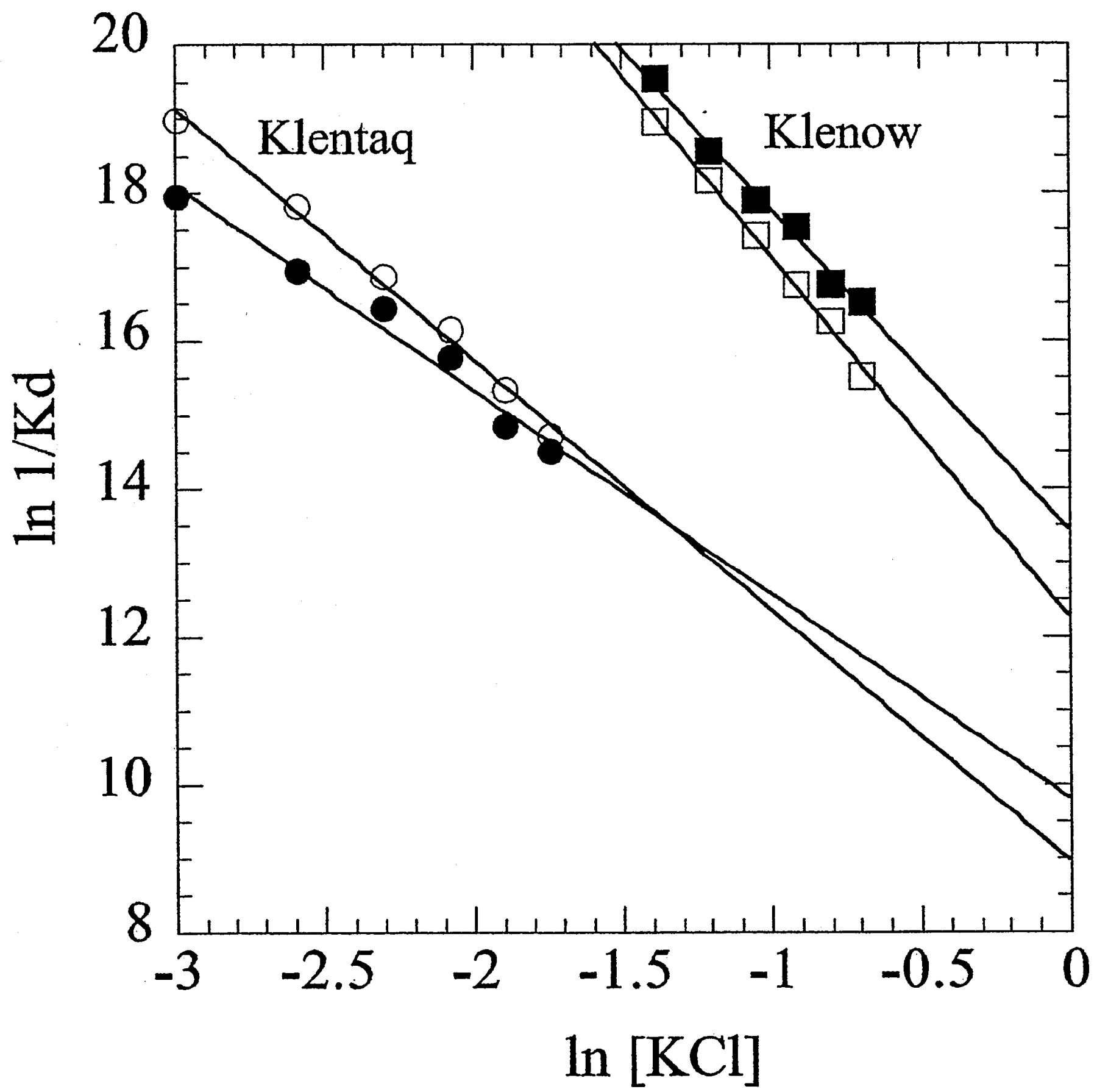
Figure 3. KCl salt linkages for primed-template DNA binding by Klenow and Klentaq. Shown are Klentaq in the presence (closed circle) and absence (open circle) of 5 mM MgCl2 and Klenow in the presence (closed circle) and absence (open circle) of 5 mM MgCl2 . The slopes of the plots give the thermodynamic net average number of ions released upon complex formation. In the presence of MgCl2, pt-DNA binding by Klenow releases 4.3 ions and Klentaq releases 2.8 ions. In the absence of MgCl2, pt-DNA binding by Klenow releases 4.9 ions and Klentaq releases 3.4 ions. (Figure is from Ref. 1)
Our salt dependence studies have thus far shown that: 1) Klenow binds 150X tighter to pt-DNA than
Taq/Klentaq, 2) The KCl “sensitivities”
and linkages (¶Ka/¶ln[KCl])
differ for the two polymerases, and 3) The MgCl2
“sensitivities” and linkages also differ for the two proteins (1). The physiological responses to salt are thus very
different for these two supposedly “homologous” enzymes.
Temperature Dependence of Binding
The DNA binding of both Klenow and Taq/Klentaq polymerases is non-linear with temperature (see Figure 4, below). This indicates that both polymerases bind DNA with large negative heat capacities (DCp). High DCp's of binding have been shown to frequently be associated with sequence specific DNA binding. Klenow and Taq/Klentaq bind DNA with little or no DNA-sequence specificity, however.
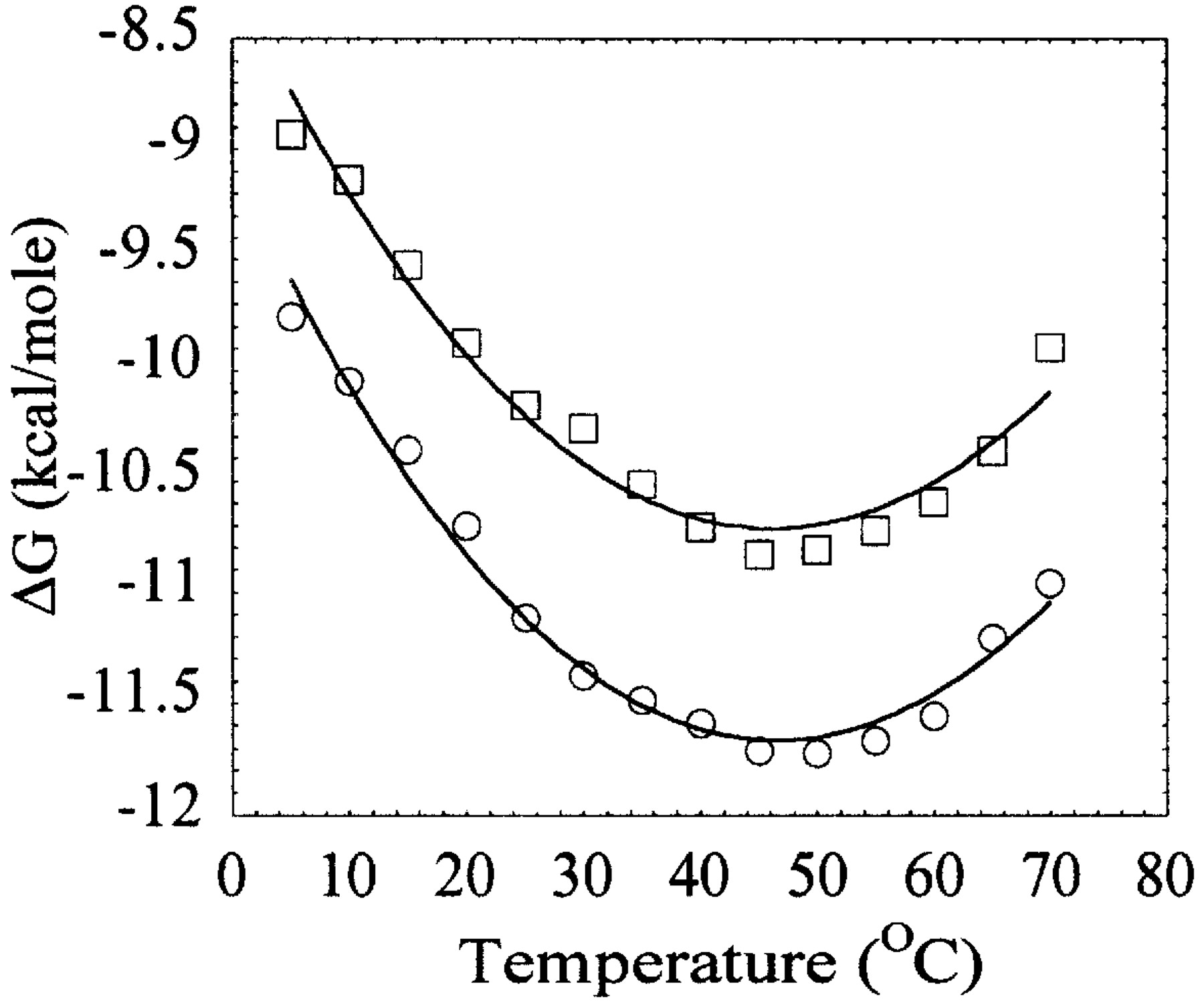
Figure 4. This plot shows the temperature dependence of the free energy (DG ) of DNA binding for Taq (circles) and Klentaq (boxes). The observed non-linearity of DG versus T results from a negative heat capacity change ( DCp) associated with the binding process, and DCp can be calculated using the Gibbs-Helmholtz equation, which allows direct calculation of the van't Hoff enthalpy ( DHvH ) and entropy ( DSvH ) as a function of temperature. The DCp for pt-DNA binding by Taq is -730 cal/mol K and Klentaq is -710 cal/mol K (figure from Ref. 2).
Klenow and Taq/Klentaq are the 3rd and 4th of four non-sequence specific DNA binding proteins that have definitively demonstrated that a zero DCp is not a “signature” of non-sequence-specific DNA binding, as had been previously postulated. Further, the commonly proposed quantitative correlation between DCp and buried surface area does not hold for Klenow or Taq/Klentaq, nor for the other two members of this DNA-binding class (2,3).
References
1. Datta K, LiCata VJ. Salt dependence of DNA binding by Thermus aquaticus and Escherichia coli DNA polymerases. J Biol Chem. 2003 Feb 21; 278(8):5694-701.
2. Datta K, LiCata VJ. Thermodynamics of the binding of Thermus aquaticus DNA polymerase to primed-template DNA. Nucleic Acids Res. 2003 Oct 1; 31(19):5590-7.
3. Datta K, Wowor AJ, Richard AJ, Licata VJ. Temperature dependence and thermodynamics of Klenow polymerase binding to primed-template DNA. Biophys J. 2006 90: 1749-1751.
4. Liu, C.-C, Richard, A.J., Datta, K., and LiCata VJ., 2008, Temperature variable heat capacity effects in protein-DNA interactions. Biophy J. 2008 94: 3258-3265.