Protein Structure in Solution
In the LiCata lab, we use techniques including small angle x-ray scattering (SAXS) and analytical ultracentrifugation (AU) to obtain information about the global size and shape of the proteins studied in our lab. We have successfully used SAXS and AU to determine the conformation of full-length Type I DNA polymerases from T. aquaticus and E. coli in an aqueous environment (1).
Global Conformations of Type I DNA polymerases in solution - Background
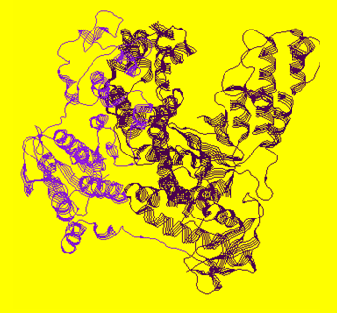
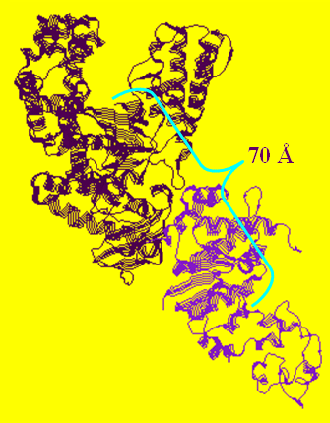
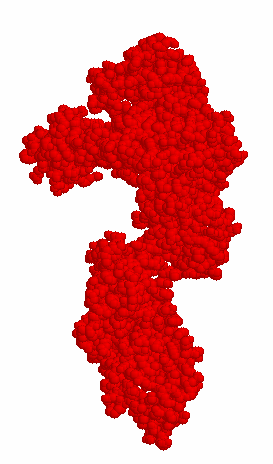
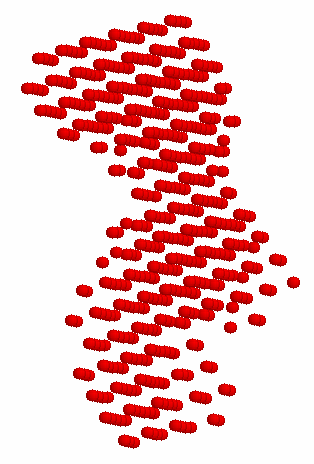
In the available crystal structures of full length apo-Taq (i.e. not ligated with a DNA substrate), the 5' nuclease domain is found positioned in two different orientations. Figure 1A shows that the 5' nuclease domain is folded up against the polymerase domain in a more compact structure, while in Figure 1B it is extended out into solution.
A (1CMW)
B (1TAQ)
Figure 1. 1CMW and 1TAQ are the Protein Data Bank (pdb) files that were used to generate these structure figures. The 5' nuclease domain of Taq DNA polymerase is either folded up against the Klentaq fragment as in 1CMW or extended out into solution (1TAQ).
The main functions of Type I DNA polymerases are replacement
of RNA primers within
SAXS - Introduction
While the SAXS technique has been utilized since the late 1930s to study non-biological polymers, it has recently gained popularity in the biological sciences. When the wavelength of electromagnetic radiation is on the same length scale as that of a sample particle, the particle will scatter the radiation. Detection and analysis of this scattering pattern can yield valuable information about the size, shape, and internal structure of the particle. The wavelength of x-rays ranges from ~0.03 to 3nm. Proteins are typically 0.1 - 10nm in diameter. Each protein particle is a scattering unit or a region of electron density. When scattering particles are randomly distributed in solution, a size, and sometimes an envelope-like model of the sample particle can be obtained from analysis of the scattering pattern.
All of our SAXS experiments are performed with the use of
synchrotron light sources. There are about 60 synchrotron light sources
worldwide and only 12 in the
|
Synchrotron facilities used by the LiCata
lab to perform SAXS experiments. |
|||
|
Ring |
Beamline |
Location |
Energy (GeV) |
|
LNLS |
D11A-SAXS |
|
~ 2 |
|
SSRL (SPEAR3) |
1-4 & 4-2 |
|
~3 |
|
Photon Factory
(PF) |
15A |
|
~3 |
|
Advanced Photon
Source (APS) |
18-ID |
|
~7 |
|
|
|
|
Figure 2. LNLS and SSRL Synchrotron pictures. |
|
|
|
|
|
Figure 3. Pictures of SAXS beamlines
at LNLS and SSRL. |
Figure 4. Graduate student, Allison J. Richard,
loading a sample on beamline D11A-SAXS at LNLS. |
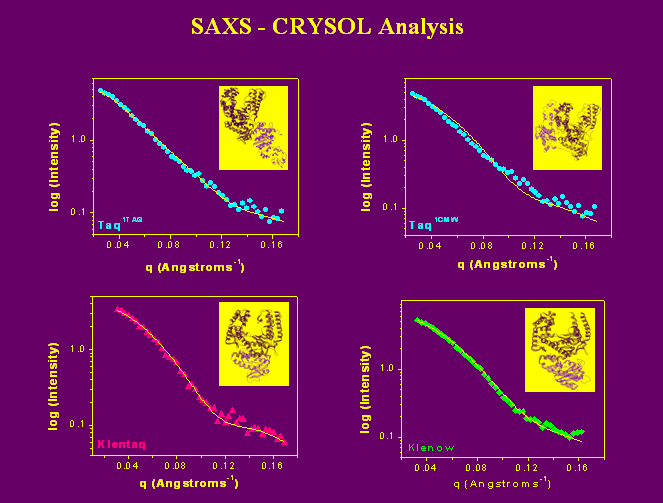
SAXS data are analyzed in a variety of different ways. Below we show the theoretical scattering curve calculated from crystal structure data (solid lines) fit to the actual experimental scattering data (3). While the predicted scattering for elongated-Taq, Klentaq, and Klenow fit the experimental data well, the fit from the predicted scattering curve of compact-Taq (top right panel) is poor.
|
|
|
Figure 5. |
More recently, we have been able to create envelope like
structural models from the scattering data alone. Figure 6 compares the modeled shape envelope
of Taq polymerase from SAXS data with the elongated Taq crystal structure, visually confirming the quantitative
shape analysis showing that full-length Taq
polymerase is in the elongated conformation in solution.
|
1TAQCrystal
Structure |
DAMMIN Model of
Taq from Experimental Scattering |
|
|
|
|
Figure 6. This figure compares the crystal
structure of Taq in the elongated conformation
(1TAQ) to the model created from the experimental scattering curve using the
DAMMIN program (4). The model provides
visual conformation to accompany the quantitative data, which indicates that
in solution the 5' nuclease domain of unligated Taq polymerase is extended away
from the Klentaq fragment. |
|
References and Useful links
1. Joubert, A.M. , Byrd, A.S., and LiCata, V.J.
(2003) Global conformations, hydrodynamics, and X-ray scattering properties of Taq and Escherichia
coli DNA polymerases in solution. J Biol Chem. 278: 25341.
2. http://www.lightsources.org/cms/?pid=1000098
- list of synchrotron facilities worldwide.
3. Svergun, D.I., Barberato, C., and Koch, M.H.J. (1995) J. Applied Crystallog. 28, 768.
4.
Svergun, D.I. (1999) Biophys. J. 76, 2879.